Xpert® MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance
Abstract
Global TB control efforts have been severely hampered by the lack of diagnostic tests that are accurate, simple to use and can be applied at the point of clinical care. This has been further compounded by the widespread inability to test for drug resistance. The Xpert® MTB/RIF assay is a rapid molecular assay that can be used close to the point of care by operators with minimal technical expertise, enabling diagnosis of TB and simultaneous assessment of rifampicin resistance to be completed within 2 h. Moreover, this can be accomplished using unprocessed sputum samples as well as clinical specimens from extrapulmonary sites. We review in detail the development of this assay, its evaluation within the laboratory, its utility among adult and pediatric TB suspects, its use as a screening tool for HIV-associated TB and studies of its implementation at the district and sub-district levels in resource-limited settings. Following endorsement by the WHO in 2010, we consider the next steps in the implementation of the assay and its potential impact in high burden settings.
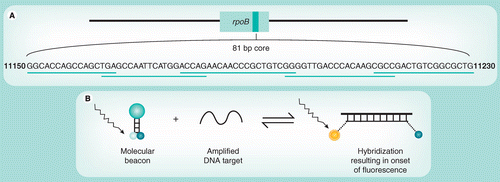
(A) The rpoB gene, the nucleotide sequence of the core region and the localization of the complementary overlapping molecular beacon probes that span the complete core region. (B) The stem-and-loop structure of a molecular beacon and onset of fluorescence following binding to a complementary DNA strand. The loop structure of the molecular beacon contains the complementary oligonucleotide probe sequence, and the fluorophore and quencher molecules are attached to the ends of the stem structure. Following hybridization, conformational change in the probe leads to separation of the fluorophore and quencher molecules and onset of fluorescence.
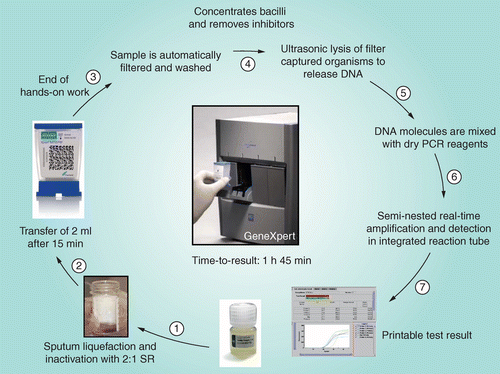
SR: Sample reagent.
Diagram supplied by C Boehme, Foundation for Innovative New Diagnostics.

Trace (A) shows the read-out from processing a rifampicin-sensitive strain of Mycobacterium tuberculosis as denoted by amplification of all five probes with a similar cycle threshold. Trace (B) shows the read-out of a rifampicin-resistant strain as shown by the failure of amplification of Probe B.
SPC: Sample processing control.

Data taken from the FIND multicountry laboratory validation study [11].

Courtesy of National Health Laboratory Service of South Africa.
The absolute number of TB cases occurring each year (9.4 million) is currently greater than at any time in history and, although the global incidence rate is estimated to have peaked and has started to decline, progress towards TB control is very slow [1]. The case detection rate worldwide in 2009 was just 63% [101] and the lack of rapid and accurate diagnostics remains a critical stumbling block, which is undermining progress towards the 2015 millennium development goals for TB control [1]. Over 90% of TB cases develop among people living in low- and middle-income countries where diagnosis still relies heavily on the use of sputum smear microscopy (a test developed over 125 years ago) and chest radiology. Despite microscopy being the diagnostic test most widely used worldwide, only 45% of TB cases that were notified in 2009 were sputum smear-positive, and these represented just 28% of the estimated total burden of incident disease globally [101].
Increasing drug resistance also poses a grave threat to TB control [2]. An estimated 440,000 cases of multidrug resistant TB (MDR-TB) occur worldwide each year and yet only 30,000 cases (approximately 7%) were diagnosed and notified to the WHO in 2009 [101]. Such low rates of case ascertainment again reflect the critical deficiency in diagnostic laboratory capacity [2,102]. Culture- or molecular-based TB drug susceptibility testing lie well beyond the scope of the vast majority of laboratories in resource-limited settings due to high cost, technical complexity and infrastructure requirements.
Growing scientific momentum in recent years, however, has fuelled a pipeline of new and improved TB diagnostics and assays to more rapidly detect drug resistance. Numbers of these assays have been endorsed by the WHO [3]. However, one of these – the Xpert® MTB/RIF assay (Cepheid Inc., Sunnyvale, CA, USA) – has been described as a potential “game changer” for TB control [4]. Using a prototype assay originally developed at the University of Medicine and Dentistry (NJ, USA) [5–7], this new TB diagnostic was developed by the Foundation for Innovative New Diagnostics (FIND) in partnership with a commercial company, Cepheid Incorporated. This has been achieved with financial support from the US NIH and the Bill and Melinda Gates Foundation. In the present article, we review the rapidly growing literature (summarized in Table 1) concerning the development of this assay and its evaluation within the laboratory and in clinical populations [5–12]. In addition we discuss unresolved issues surrounding the implementation of this assay in resource-limited settings and the many remaining research questions.
Assay development
Nucleic acid amplification tests have long held great promise for TB diagnosis and the rapid detection of drug resistance, and commercial assays have been widely used within high-income countries for over 20 years [3]. However, despite high specificity, sensitivity for TB diagnosis has been modest and variable in sputum smear-negative and extrapulmonary TB [13–16]. The highest rates of smear-negative TB occur among HIV-infected patients, and 80% of the global burden of HIV-associated TB lies in sub-Saharan Africa [17] where laboratory diagnostic capacity is least well developed.
The WHO took the bold step of endorsing nucleic acid amplification tests for use in resource-limited settings for the first time in 2008 to rise to the challenge of global drug resistance epidemic. With the growing evidence of the true scale of the MDR-TB epidemic, molecular line probe assays were endorsed for rapid detection of drug-resistant TB [103]. However, these assays are technically demanding, costly, use is restricted to centralized laboratories, and they are not recommended for directly testing smear-negative clinical specimens in view of limited sensitivity and risk of cross-contamination. Scale-up and impact of these assays has therefore been limited.
The newly developed Xpert MTB/RIF assay overcomes a number of these limitations. It utilizes real-time PCR (rt-PCR) technology to both diagnose TB and detect rifampicin resistance concurrently using unprocessed clinical specimens, regardless of their smear status. The assay is conducted within a simple, almost fully automated cartridge-based system. The simplicity for the user makes this an assay that could feasibly be widely implemented outside centralized laboratories and potentially impact on TB control.
Target DNA sequences
Rifampicin resistance is particularly amenable to rapid molecular detection since >95% of all rifampicin resistant strains contain mutations localized within the 81 bp core region of the bacterial RNA polymerase rpoB gene, which encodes the active site of the enzyme [18]. Moreover, mutations that occur in this region are highly predictive of rifampicin resistance, whereas susceptible isolates almost always have the same wild-type nucleotide sequence [18,19]. In addition, the rpoB core region is flanked by Mycobacterium tuberculosis-specific DNA sequences. Thus, it is possible to test for M. tuberculosis and for rifampicin resistance simultaneously, by targeting a single amplicon generated using PCR technology. Moreover, rifampicin resistance is strongly, although not invariably, indicative of MDR-TB (defined by concomitant resistance to isoniazid – another key antituberculosis agent). Resistance to isoniazid, by contrast, is conferred by mutations in a number of genes and is a poor marker of MDR-TB since isoniazid monoresistance is common. By targeting the two most common of these genes, katG and inhA, using a prototype assay to test clinical isolates from Spain and the USA, the sensitivity for detection of isoniazid resistance was only 85% compared with 98% for detection of rifampicin resistance by targeting the rpoB gene [6]. Thus, the rpoB gene represents a much better molecular target for the simultaneous detection of TB and the key form of drug resistance.
Molecular beacon technology
The Xpert MTB/RIF assay utilizes molecular beacon technology [20,21] to detect DNA sequences amplified in a hemi-nested rt-PCR assay. Five different nucleic acid hybridization probes are used in the same multiplex reaction [5,6]. Each probe is complementary to a different target sequence within the rpoB gene of rifampicin-susceptible M. tuberculosis and is labeled with a differently colored fluorophore. Together, these overlapping probes span the entire 81 bp core region of the rpoB gene (Figure 1A)[5,6].
The molecular beacons are oligonucleotide sequences that contain a probe sequence inserted between two ‘arm’ sequences. The two ‘arm’ sequences are designed to be complementary to each other such that, under assay conditions, they hybridize to form a stem-and-loop secondary structure (Figure 1B); the probe is located within the loop structure [20]. A fluorophore is covalently linked to the end of one arm and a nonfluorescent quencher to the other. With the probe in its free, nonhybridized state, the close proximity of the quencher and fluorophore molecules supresses fluorescence. However, when the probe sequence binds to its complementary DNA target, the molecular beacon undergoes conformational change. This causes separation of the two arms and the fluorophore and quencher molecules, resulting in the onset of bright fluorescence [20].
The molecular beacons were designed to only hybridize correctly with amplified wild-type (rifampicin sensitive) rpoB sequences [5,6]. A mutation within these sequences interferes with hybridization such that the conformational integrity of the probe may be retained in the nonfluorescing state. Thus, a mutation anywhere in the core region of the rpoB gene results in either delayed onset (partial inhibition) or complete suppression of fluorescence of the corresponding molecular beacon [5]. Using genomic DNA or culture lysates of a large number of clinical strains of M. tuberculosis, this prototype assay was found to have high sensitivity and specificity for detection of rifampicin resistance [5–7]. A further important step forward was the development of a version of the assay that could successfully be performed in a single well [7].
The GeneXpert® diagnostic platform
The GeneXpert diagnostic system was originally developed by Cepheid Inc. for the detection of anthrax [22] and was deployed for this purpose by the United States Postal Service in mail sorting facilities. This application required the development of a self-contained, fully integrated and automated platform that could be operated with minimal technical expertise [104]. The GeneXpert system has been successful in combining on-board sample preparation with fully-automated rt-PCR amplification and detection functions. The cartridge-based system incorporates microfluidics technology and fully automated nucleic acid analysis to purify, concentrate, detect and identify targeted nucleic acid sequences from unprocessed clinical samples. An expanding range of different organisms may be detected using pathogen-specific cartridges within the same GeneXpert test platform. The test platform is modular, with each module independently processing one cartridge at a time. Machines with 1, 4, 16 and 48 modules are available, permitting multiple assays to be run concurrently and independently [104].
The Xpert MTB/RIF assay
The assay utilizes single-use plastic cartridges with multiple chambers that are preloaded with liquid buffers and lyophilized reagent beads necessary for sample processing, DNA extraction and hemi-nested rt-PCR [8,9]. Clinical sputum samples (or decontaminated sputum pellets) are treated with a sodium hydroxide and isopropanol-containing sample reagent (SR). The SR is added to the sample (currently recommended at a 3:1 ratio for sputum pellets and a 2:1 ratio for unprocessed sputum samples) and incubated at room temperature for 15 min. This step is designed to reduce the viability of M. tuberculosis in sputum at least 106-fold to reduce biohazard risk [10]. The treated sample is then manually transferred to the cartridge which is loaded into the GeneXpert instrument. Subsequent processing is fully automated. A schematic of the assay procedure is shown in Figure 2.
The cartridge incorporates a syringe drive, a rotary drive and a filter upon which M. tuberculosis bacilli are deposited after being liberated from the clinical material. The test platform employs a sonic horn that inserts into the cartridge base to cause ultrasonic lysis of the bacilli and release of the genetic material. The assay then amplifies a 192 bp segment of the rpoB gene using a hemi-nested rt-PCR reaction [8,9]. The assay also contains lyophilized Bacillus globigii spores which serve as an internal sample processing and PCR control. The B. globigii PCR assay is multiplexed with the M. tuberculosis assay.
Mycobacterium tuberculosis is detected by the five overlapping molecular probes (probes A–E) that collectively are complementary to the entire 81 bp rpoB core region [8,9]. M. tuberculosis is identified when at least two of the five probes give positive signals with a cycle threshold (CT) of ≤38 cycles and that differ by no more than a prespecified number of cycles. The B. globigii internal control is positive when the single B. globigii-specific probe produces a CT of ≤38 cycles [8,9]. The standard user interface indicates the presence or absence of M. tuberculosis and the presence or absence of rifampicin resistance, and a semi-quantitative estimate of the concentration of bacilli as defined by the CT range (high, <16; medium, 16–22; low, 22–28; very low, >28). Assays that are negative for M. tuberculosis and for the B. globigii internal control are reported as invalid assays. When performed on unprocessed sputum samples, the assay can generate results within 2 h with less than 15 min of hands-on time.
The basis for detection of rifampicin resistance is the difference between the first (early CT) and the last (late CT) M. tuberculosis-specific beacon (ΔCT). The system was originally configured such that resistance was reported when ΔCT was >3.5 cycles and sensitive if ≤3.5 cycles. Since the assay terminates after 38 cycles, the assay was deemed indeterminate for rifampicin resistance if the first probe CT is >34.5 cycles and the last probe has a CT of >38 cycles [8,9]. From May 2010 the automated detection of rifampicin resistance was modified using a new ΔCT cut-off in order to improve the specificity for rifampicin resistance detection (see later) [12]. Figure 3 shows traces from rifampicin-susceptible and rifampicin-resistant strains of M. tuberculosis identified in sputum samples from Cape Town.
Laboratory evaluations
Analytic performance: sensitivity
In the first report of the Xpert MTB/RIF assay, the limits of detection of the assay were assessed [8]. By adding M. tuberculosis genomic DNA directly into the cartridge, the limit of detection (95% sensitivity) was found to be 4.5 (95% CI: 3.3–9.7) genome copies. Clinical sputum samples from non-TB patients were also spiked with known numbers of M. tuberculosisH37Rv bacilli (10–300 CFU per ml of sputum) and multiple replicates tested. The limit of detection was found to be 131 (95% CI: 106–176) CFU/ml of sputum [8]. This compares with the limits of detection of automated mycobacterial liquid culture, which lie in the range of 10–100 CFU/ml, compared with an estimated 10,000 CFU/ml for smear microscopy [23]. Thus, the MTB/RIF assay has a sensitivity that is approximately two orders of magnitude greater than smear microscopy and that approaches that of culture.
The dynamic range of the assay was assessed by spiking 102 –107 CFU of M. tuberculosis H37Rv into sputum samples and a log-linear relationship with cycle threshold was observed throughout this range with no impact on amplification of the internal control [9]. Assay sensitivity was further assessed by testing low concentrations of genomic DNA from 79 phylogenetically and geographically diverse strains of M. tuberculosis, all of which were correctly identified as M. tuberculosis[9].
Analytic performance: specificity
In an initial assessment of assay specificity, 20 different isolates of nontuberculous mycobacteria (NTM) (including 16 species that commonly cause human disease) were spiked into unprocessed sputum samples [8]. All tested negative, showing high assay specificity for M. tuberculosis. Moreover, experiments in which high concentrations of NTM bacilli were mixed with low concentrations of M. tuberculosis in sputum samples showed no impairment of assay sensitivity. In a further assessment, the assay was tested with a panel of 89 different organisms, including bacteria, fungi and viruses, using strains commonly found in the respiratory tract [9]. Each test was done with 106 genomes of the isolate and all tested negative (100% specificity). Two of the assay probes amplified with Nocardia asteroides, but the ΔCT value was consistently greater than two cycles and, therefore, was not reported as M. tuberculosis. Further experiments have examined the potential impact of assay cross-contamination with amplicons from the MTBDRplus assay (Hain LifeSciences, Nehren, Germany) [9]. False-positive results were not generated until at least 108 copies of the amplicon were spiked into 1 ml of sputum, suggesting that false-positive results would be very unlikely to occur from this source.
Analytic performance: rifampicin resistance detection
Genomic DNA from M. tuberculosis isolates with 23 different commonly occurring rpoB mutations were tested and all were detected by Xpert MTB/RIF (100% sensitivity) [8]. A total of 16 of the 23 mutations caused complete failure of detection of at least one of the five probes and the remaining mutations produced a ΔCT of >3.5 cycles. All DNA samples from rifampicin susceptible isolates were correctly identified as susceptible with an average ΔCT of 1.8 cycles [8]. Assay sensitivity and specificity for rifampicin detection was further assessed by testing low concentrations of genomic DNA from 79 phylogenetically and geographically diverse strains of M. tuberculosis that included 42 rifampicin susceptible and 37 resistant strains [9]. Again, sensitivity and specificity for detection of rifampicin resistance were both 100%.
Further experiments were done in which DNA from resistant and susceptible strains was mixed in varying ratios to assess how this impacted detection of rifampicin resistance [9]. To enable detection, between 65% and 100% of the DNA from the rifampicin-resistant isolate had to be present, depending on the mutation [9]. Whereas some mutations completely block probe hybridization, others only inhibit it. If only partial inhibition occurs, then the presence of only a small concentration of wild-type amplicon would need to be present to boost the probe signal into the normal range. Overall, this suggests that in patients with mixed infections, the Xpert MTB/RIF assay might only detect the resistant strain if it is the predominant one present. However, selection of resistant strains during the course of standard TB treatment might lead to an apparent switch from a susceptible to a resistant phenotype when comparing baseline testing with repeat testing during treatment.
Assessment of biosafety
For the Xpert MTB/RIF assay to be used outside the laboratory environment, demonstration of its capacity for biohazard containment was essential. Experiments were done to assess the sterilizing capacity of the SR and to compare bioaerosol generation by the Xpert MTB/RIF assay with direct smear microscopy [10].
Killing studies demonstrated that the viability of M. tuberculosis was reduced by over 8 logs within 15 min of incubation in SR and that the cycle threshold was unaffected by incubation in the reagent for up to at least 24 h [8]. However, very prolonged incubation in SR for 3 days can cause false positive rifampicin resistance results due to chemically induced deamination causing mutations in the rpoB region. In further experiments, sputum samples (n = 45) with high smear positivity grading from patients with TB were divided into three and treated with one of two concentrations of SR (3:1 or 2:1 ratio of SR to sputum) or with standard N-acetyl-cysteine/sodium hydroxide treatment (positive control) and then cultured [10]. None of samples treated with SR (3:1) were culture-positive whereas growth occurred from 3/45 samples treated with SR (2:1), albeit with a much longer time to positivity compared with the positive controls. Storage of SR at a range of temperatures between 4°C and 45°C for 3 months did not affect sterilizing capacity [10].
Bioaerosol generation was assessed by spiking 5 × 108 CFU/ml of BCG into sputum samples that were then tested with Xpert MTB/RIF assay or by standard smear microscopy [10]. During testing procedures, bioaerosol generation was assessed in a variety of laboratory conditions using two different air sampling devices. Viable bioaerosols were not generated during the manual sputum processing with SR or during automated processing, even when cartridges were directly loaded with high concentrations of viable organisms [10]. In contrast, viable airborne organisms were detected during routine preparation of smears. These data therefore suggested that the use of the Xpert MTB/RIF assay poses a substantially smaller biohazard risk than direct smear microscopy and might reasonably be done without the need for special biosafety equipment, which is lacking in most resource-limited settings.
Evaluation in clinical populations
An early small-scale clinical validation study of Xpert MTB/RIF using archived frozen sputum samples produced very encouraging results [8] and paved the way for much larger evaluations in clinical populations.
Multicountry evaluation study by FIND
The seminal evaluation of the Xpert MTB/RIF assay was designed and supervised by the FIND [11]. The study was designed to assess diagnostic accuracy rather than impact of using the assay as near-patient technology on patient outcomes. Thus, all assays were conducted in centralized laboratories. The study sites (n = 5) in Peru, Azerbaijan, South Africa (two sites), and India were chosen to represent a diversity in the prevalence of TB, HIV coinfection and multidrug resistance. Symptomatic patients (n = 1730) with chronic cough who were suspected of having drug-sensitive or drug-resistant TB and who were able to produce three 1.5 ml volume sputum samples were enrolled. Overall, 40.2% of patients enrolled had HIV infection and 46.4% had a history of previous TB treatment [11].
The three sputum samples obtained from each patient were randomly allocated either to be directly tested with microscopy and the Xpert MTB/RIF assay (one sample) or to be processed by standard decontamination with sodium hydroxide and N-acetyl-cysteine and centrifugation with subsequent microscopy and culture using solid and liquid media (two samples). TB cases were defined by one or more positive cultures.
A single direct test on sputum detected 551 of 561 (98.2%) patients with smear-positive TB and 124 of 171 (72.5%) patients with smear-negative TB [11]. The test was specific in 604 of 609 (99.2%) patients without TB, as defined using a combined reference standard of culture and clinical review. Among patients with smear-negative TB, processing one, two or three samples was associated with sensitivities of 72.5%, 85.1% and 90.2%, respectively (Figure 4).
Compared with the results of phenotypic drug susceptibility testing, the assay also correctly identified 200 of 205 patients (97.6%) with rifampicin-resistant isolates and 504 of 514 patients (98.1%) with rifampicin-susceptible isolates [11]. Among the 15 patients with discordant rifampicin resistance results, sequencing of the rpoB gene found mutations in 9 of 10 isolates in which the Xpert MTB/RIF assay results appeared to be false-resistant results; in addition, one apparently false-sensitive Xpert MTB/RIF result was confirmed as wild-type by sequencing and three other patients had mixed infections. Thus, all but two of the discrepant results were resolved, and taking these revised data into account, the sensitivity and specificity for rifampicin resistance were recalculated as being 99.1% and 100%, respectively. The 16 rpoB mutations detected in this study were broadly representative of prevailing resistance mutations globally.
The Xpert assay was indeterminate in 3.7% of tests performed, which was lower than the rate of culture contamination (5.5%) [11]. The indeterminate rate dropped to 1.2% with the inclusion of the results from a second sample. Among samples in which M. tuberculosis was detected by Xpert, 1.0% had an indeterminate result for rifampicin resistance. All these samples were smear-negative with a high cycle threshold (≥35 cycles).
Assay sensitivity was not associated with whether the sputum sample was directly tested or tested following decontamination and centrifugation [11]. Moreover, the results of this study were similar across all study sites, indicating that the findings are likely to be broadly applicable. However, a culture-confirmed diagnosis of TB was made among 38.8% of patients overall (substantially higher than among most routine populations of TB suspects), suggesting considerable patient preselection.
The remarkable results of this first large-scale evaluation propelled the Xpert MTB/RIF assay to the center stage in the TB field and provided the main evidence supporting the WHO endorsement of the assay in December 2010, discussed later.
Further studies of diagnostic accuracy in patients with suspected TB
A total of 12 studies conducted in the USA, Europe, India and sub-Saharan Africa have assessed the utility of Xpert MTB/RIF assay in clinical populations of patients suspected of having TB (Table 1). These are largely laboratory-based studies of respiratory and nonrespiratory clinical samples. However, several studies of prospectively enrolled cohorts are now emerging.
Respiratory samples
Four laboratory-based studies conducted in the USA [24], France [25], Spain [26] and The Netherlands [27] tested routine respiratory samples and found a very similar performance as the FIND multicountry evaluation [11]. Sensitivities for smear-positive TB were 98–100% [24,25], and ranged between 57 and 83% for smear negative pulmonary TB [24–26]. Sensitivity was inversely related with time to positivity in culture but was not related to duration of frozen storage (range: 1–10 years) in the study by Moure et al.[26]. Assessment of specificity in laboratory-based studies is limited, since obtaining further samples and clinical follow-up is often not possible. However, four false-positive Xpert results reported by Marlowe et al. were all thought to be true positives, three being from patients already receiving TB treatment and one being from a patient later proven to have culture-positive disease [24]. Moreover, there was no cross-reactivity with 41 NTM isolates.
A single study from South Africa has reported on the utility of Xpert in pediatric in-patients suspected of having TB [28]. Diagnosis of pediatric TB is notoriously difficult and often lacks laboratory confirmation [29]. Induced sputum specimens were obtained from 452 children with a median age of 19.4 months. Using liquid culture as the reference standard, two Xpert tests doubled the case detection rate compared with smear microscopy (76% vs 38%), identifying all smear-positive and 61% of smear-negative cases; specificity was 98.8% [28]. The sensitivities for smear-negative TB were 33.3% and 61.1% when testing one or two samples, respectively. Xpert diagnosed TB after a median of 1 day compared to 12 days using culture. The study strongly supports a policy of testing two induced sputum samples using Xpert from pediatric TB suspects [28].
Two additional studies aimed to assess the diagnostic accuracy of the Xpert MTB/RIF assay among adult TB suspects by testing frozen sputum samples collected during earlier studies, but the information derived from these is limited by study design [30,105]. Theron and colleagues in South Africa collected two spot sputum samples, one of which was examined by smear microscopy and cultured in liquid media at the time of sample collection, while the second was frozen and retrospectively tested with the Xpert MTB/RIF assay [30]. Analysis included 480 patients in whom 141 had culture-confirmed TB. Sensitivity was 94.7% for smear-positive TB and 46.8% for smear-negative TB, with an overall specificity of 78.7%. The study was limited by the fact that only one of the sputum samples was cultured and it cannot be assumed that the bacillary concentrations in the paired samples tested with Xpert were the same as those in the cultured sample, potentially giving rise to apparent discordance. Post-hoc analysis that added a diagnostic category of ‘probable TB’ using other diagnostic criteria increased the calculated specificity but not the overall sensitivity of the Xpert assay [30].
Xpert has also been evaluated using frozen sputum samples collected during a previous TB diagnosis study in Tanzania [105]. TB suspects (n = 292) were studied yielding 61 culture-positive cases. While the study confirmed high sensitivity for smear-positive disease (98%) and high specificity among those with NTM cultured, the study was limited by the small number of patients with smear-negative culture-positive TB and by 77 patients being categorized as having ‘clinical TB’ with no microbiological confirmation [105].
Nonrespiratory samples
Laboratory-based studies in Germany, France and Spain have assessed the utility of Xpert for TB diagnosis in nonrespiratory samples, including pleural fluid, gastric fluid, tissue, cerebrospinal fluid, urine and stool [25,31,32]. Hilleman and colleagues tested 521 samples and found an overall sensitivity of 77.3% and a specificity of 98.2%, which is comparable with results from studies of smear-negative respiratory samples [31]. Causse and colleagues tested a wide range of nonrespiratory samples (n = 341) and reported a higher sensitivity of 95% and specificity of 100%, outperforming the commercially available Cobas TaqMan® MTB assay [32]. By contrast, the study by Armand and colleagues included results from just 32 nonrespiratory samples and found an overall sensitivity of 53% that was substantially lower than that obtained using an in-house IS6110-based real-time PCR assay (78%) [25].
Excellent results were also reported in a study from India in which patients (n = 546) with suspected extrapulmonary TB at diverse anatomic sites were prospectively enrolled and studied [33]. A reference standard of smear, culture, radiology, histology and clinical findings was used. The overall sensitivity and specificity were reported as being 81 and 99.6%, respectively. Similar to studies of pulmonary TB, sensitivity was higher for smear-positive specimens (96%) compared with smear-negative specimens (64%) [33].
A further preliminary report from South Africa described the utility of Xpert MTB/RIF for increasing case detection among HIV-infected TB suspects (n = 101) routinely referred for diagnostic lymph node fine needle aspiration. Use of the assay to test remaining clinical material from these aspirates was associated with a substantial increase in case detection compared to smear microscopy and considerably reduced the time to diagnosis [34].
Collectively, these results indicate substantial utility of Xpert MTB/RIF for diagnosis of extrapulmonary TB and further prospective evaluations of clinical suspects are warranted to more precisely define the utility for different forms of extrapulmonary disease.
Screening for HIV-associated TB
A single study has evaluated the utility of Xpert MTB/RIF for screening for HIV-associated TB among patients with advanced immunodeficiency prior to starting antiretroviral therapy (ART) in a South African township [35]. In this clinical setting, the high burden of TB is a major cause of morbidity, mortality and nosocomial disease transmission [36]. However, TB diagnosis is also extremely challenging since symptom screening, chest radiology and smear microscopy all perform poorly and culture-based diagnosis often takes many weeks [36–39].
In a prospective study, a cohort of 515 patients were screened for TB regardless of the presence or absence of symptoms by obtaining two sputum samples [35]. The prevalence of TB was just under 20% using liquid culture as the gold standard. Xpert increased the case detection by 45% compared to smear microscopy (73% vs 28%) with a specificity of 99.2%. When testing one or two sputum samples, the overall sensitivities of Xpert were 58% and 72%, respectively, and were 43% and 62% for sputum smear-negative disease [35]. Low sensitivity for smear-negative disease was related to an absence of cough of at least 2 weeks duration and prolonged time to positivity in culture (reflecting low organism load). The assay also correctly identified four cases of MDR-TB, reducing median time to diagnosis by over 5 weeks and showing the great potential of the assay to reduce the risks of nosocomial MDR-TB outbreaks in HIV care and treatment settings. However, there were also three confirmed false-positive rifampicin resistance results (see the next section) [35].
This study also simulated the utility of the assay in a hypothetical cohort of HIV-infected patients incorporating the assay in a range of different screening algorithms [35]. In settings with high TB prevalence such as in South Africa, there was a strong rationale for screening all patients with either one (or ideally two) Xpert tests regardless of symptoms. However, at low TB prevalence rates, this would be less likely to be a cost-effective strategy.
False-positive rifampicin resistance results
Despite the findings of extremely high specificity for detection of rifampicin resistance in the initial FIND evaluation study [11], several subsequent studies have detected confirmed false-positive rifampicin resistance results. In a study from the USA [24], all 130 culture-confirmed TB isolates were rifampicin-susceptible by conventional testing, but three were reported as resistant by Xpert. Repeat Xpert testing or rpoB gene sequencing confirmed that the initial results were false-positives and all three showed a delay in the CT for probe B. Similar false-positive results confirmed by rpoB gene sequencing have also been reported from studies in South Africa, including the FIND implementation study [35,106].
Although absolute numbers of such cases have been quite small, in sites with a low prevalence of rifampicin resistant isolates, this results in a low positive predictive value for a result reporting rifampicin resistance, which is very problematic for clinical decision making [106]. In a TB screening study from South Africa, the sensitivity for detecting rifampicin resistance was 100%, but several additional false-positive results meant that the positive predictive value was just 57% [35]. The utility of the assay for diagnosing rifampicin resistance is strongly related to the prevalence of MDR-TB. WHO estimates that the positive predictive value is over 90% where this exceeds 15% of isolates, but is less than 70% when the prevalence is under 5% [107].
If Xpert MTB/RIF were used to directly guide patient management, then some patients could inappropriately receive an MDR-TB diagnosis resulting in a treatment regimen with reduced efficacy (no rifampicin or isoniazid), high toxicity, prolonged duration, marked stigmatization and prolonged hospitalization. Thus, WHO recommends that when rifampicin resistance is detected by Xpert, further culture-based investigation and drug susceptibility testing for first- and second-line drugs should be done. The patient should receive an MDR-TB treatment regimen pending further results [107].
With the discovery of numbers of sequence-confirmed false-positive rifampicin resistance results during the FIND implementation study (described later) [12], the software cut-off that defines drug resistance was redefined in May 2010, although false-positive results have persisted despite this [35]. The authors also understand that further research and development work has been done to more clearly define this problem, leading to reconfiguration of the software controlling the assay process and redesign of one of the molecular probes. However, no data has been published on these changes. This problem with the assay is a critical issue that requires rapid resolution in view of moves towards large-scale implementation.
Multicountry decentralized implementation study by FIND
All studies reviewed thus far were conducted with the GeneXpert diagnostic platform located within centralized laboratory facilities. The next critical step in the evaluation of this assay was to examine its utility in district and sub-district health facilities in countries with high TB burden and to evaluate its impact on patient treatment initiation and outcomes. FIND conducted a multicenter evaluation in urban health centers in South Africa, Peru and India, drug resistance screening facilities in Azerbaijan and the Philippines, and an emergency room in Uganda [12]. In all centers, the GeneXpert machines were located within laboratories at the health facility where smear microscopy was being performed.
Adult TB suspects (n = 6648) with cough >2 weeks were investigated, comparing the utility of Xpert MTB/RIF with microscopy performed within the health facility compared to a gold standard of sputum culture carried out in centralized laboratories [12]. Testing a single sputum sample with the Xpert MTB/RIF test detected 933 of 1033 (90.3%) culture-confirmed TB cases compared with a sensitivity of 67.1% using smear microscopy. The sensitivities of Xpert were 98.3% for smear-positive cases and 76.9% for smear-negative cases, and overall specificity was 99.0%. Overall sensitivity in HIV-infected patients tended to be lower than that observed in non-HIV-infected patients (82.4 vs 90.7%, respectively) although this did not reach statistical significance (p = 0.085). The rate of indeterminate results from Xpert assays was 2.4% compared to 4.6% of cultures.
The Xpert MTB/RIF assay greatly accelerated the time to diagnosis, with a median time of 0 days compared to 1 day for smear microscopy, 16 days using liquid culture and 20 days using solid culture [12]. For patients with smear-negative TB, the Xpert assay reduced the time to start of treatment from 56 days (interquartile range [IQR] 39–81) to 5 days (IQR, 2–8). Rates of untreated smear-negative culture-positive TB decreased from 39.3% without Xpert to 14.7% using the assay to direct treatment initiation. The study did not, however, ascertain outcomes of TB treatment, and any impact on TB transmission remains unknown.
Test sensitivity for rifampicin resistance was 96.8% (242 of 250) and specificity was 96.2% (779 of 810) [12]. With the discovery of numbers of sequence-confirmed false-positive rifampicin resistance results, the software cut-off that defines drug resistance was redefined in May 2010. Using this revised cut-off in a post-hoc analysis, the specificity increased to 98.3% but the sensitivity decreased to 94.4%. The time to detection of rifampicin resistance was 1 day (IQR, 0–1) using the Xpert test, 20 days (10–26) using a line-probe assay (done on sputum pellets for smear-positive samples and culture isolates for smear-negative specimens), and 106 days (IQR, 30–124) for phenotypic drug susceptibility testing. Moreover, the likelihood of resistance results reaching the clinic was far greater compared to results of the assays conducted in centralized laboratories. However, the Xpert assay resistance results were not used to inform treatment regimen decisions in this study and the impact on outcomes of treatment for drug-resistant TB are unknown.
The GeneXpert platform was found to be easy to use, with operators with no molecular biology laboratory experience or computer skills being proficient within 1–3 days of training (Figure 5). No contamination was detected during monthly negative control runs. There were two problems with the test platform, with one requiring replacement of the machine and the other requiring replacement of a single module. All sites had power interruptions, but successfully used uninterruptible power supplies to maintain machine usage.
This study demonstrating the ability to rapidly and reliably detect 90% of TB cases including nearly 77% of smear-negative cases is a major breakthrough, and this is the first study to demonstrate the major impact on time to diagnosis and start of treatment.
WHO endorsement & implementation
In the latter part of 2010, WHO reviewed all the available evidence regarding the Xpert MTB/RIF assay, initially through an expert group, then through the Strategic Technical Advisory Group for TB (STAG-TB) and finally within a wider WHO consultation process. In December 2010 WHO endorsed the use of the assay [108] as follows:
▪ Strong recommendation: the Xpert MTB/RIF assay should be used as the initial diagnostic test in individuals suspected of having MDR-TB or HIV-associated TB;
▪ Conditional recommendation: recognizing the major resource implications, the Xpert MTB/RIF assay may be used as a follow-on test to microscopy in settings where MDR-TB and / or HIV are of lesser concern, especially in smear-negative specimens.
The WHO roadmap accompanying this endorsement recommended an initial phased implementation of the assay from 2011 onwards, leading to more rapid scale-up thereafter [109]. This may be done at a national level by individual countries (such as South Africa and India, which have developed country plans) and this might be facilitated by assistance from a range of agencies and initiatives such as PEPFAR, TB CARE, EXPAND-TB, TB REACH, The World Bank and The Global Fund. Guidelines for implementation have subsequently been issued [107] but these will need to be rapidly updated as experience and new data are gained. An important issue is that implementation of this assay will also require the operational infrastructure to ensure that detection of rifampicin-resistant isolates by Xpert can be confirmed by further drug susceptibility testing. Moreover, it is essential that there is sufficient capacity to adequately treat all the cases of MDR-TB that are diagnosed.
Costs & maintenance
A major issue facing low- and middle-income countries wishing to implement this technology is the high cost compared to smear microscopy. However, FIND has negotiated substantial cost reductions (relative to prices in the USA and Europe) for use in the public sector in 116 high burden low- and middle-income countries [109] The initial capital cost for the GeneXpert instrument in these settings (approximately US$17,500 per 4-module instrument) is considerably more than for microscopy (approximately $1,500 per light emitting diode [LED] microscope) but much lower than for conventional culture and DST, especially in view of the savings from dispensing with the need for expensive biosafety equipment [109]. Instruments require annual calibration, which is likely to prove difficult in remote settings, but the cost of this may be minimized by the proposed development of a web-based system.
The average cost per cartridge in Europe is €50. However, the discounted cost per cartridge in low- and middle-income countries has been projected to fall over time according to global volumes used (Table 2). The cost per test in early 2011 (estimated at $18 per test) is substantially greater than for microscopy, though similar to costs for performing culture and drug susceptibility testing (approximately $20 on solid media and $30 on liquid media). In the short-term, implementation of the assay would represent a substantial financial increase in diagnostic costs for national ministries of health but if the full benefits of the assay in terms of clinical outcomes and TB control are favorable, this may prove to be a wise investment.
Unresolved issues
While much has rapidly been learned about this assay, much further research is needed in the following four main categories:
Further diagnostic applications of the assay
Pediatric populations
Only a single study in pediatric population has been published [28] and further data are needed in different settings and age groups.
Use for routine screening high-risk populations
Studies to date have almost exclusively examined diagnostic accuracy in symptomatic TB suspects. Only one study has examined the utility of the assay as a screening test and that was done among HIV-infected patients preparing to start antiretroviral therapy [35]. Further screening studies in high-risk populations are needed as well studies to define how Xpert best fits into screening algorithms.
Extrapulmonary TB
Data accrued thus far on extrapulmonary TB are few and have largely been opportunistic laboratory-based studies using routine samples obtained from a variety of anatomic sites. Systematic studies of various forms of extrapulmonary TB are now needed, including investigation of requirements for optimum sample preparation.
Use for monitoring response to TB treatment
The utility of the assay for monitoring response to TB treatment (drug-sensitive and drug-resistant disease) or for detecting the emergence of drug-resistant disease is not known. The assay requires the presence of intact organisms in the sample, as these are deposited on the cartridge filter during the initial automated steps of the assay. It is not known how long intact but nonviable mycobacteria persist during TB treatment, but detection of these might cause false-positive results.
Further research & development for the rifampicin resistance read-out
As discussed above, reports of confirmed false-positive rifampicin resistance have emerged from recent studies and further research and development is urgently needed to develop ‘fixes’ for this and then to validate these in the field. Interim solutions also need to be identified to know how to optimally use the current form of the assay (see earlier).
Operational implementation
Following endorsement by the WHO in December 2010, the green light has been given for implementation in resource-limited settings. There are many questions regarding how to scale up use of this assay, including within which levels of the health service to use it; whether this is viable in rural settings or even mobile units; whether this platform can be run from a battery power supply in areas with unreliable electricity supplies; how durable the hardware is; how annual calibration of the instrument can be simplified. Secondly, research needs to clearly define where the assay is incorporated in the diagnostic algorithm for TB suspects in the context of limited resources; whether this should replace microscopy or be used as an ‘add-on’ test; whether use of two cartridges is justified in some situations; how to manage patients with negative Xpert results. In addition, optimized management of patients with positive rifampicin resistance results needs to be carefully defined. In some settings, such as parts of South Africa, for example, rifampicin resistance is less highly predictive of MDR-TB, since high rates of rifampicin mono-resistance also occur [40]. Such patients should not be inadvertently denied the benefits of isoniazid therapy. It also needs to be defined how the TB programme should best adapt to use this technology effectively and how notification and reporting systems (traditionally based on sputum smear status) need to be modified.
Impact assessment
This assay is expensive, and strong justification for scale-up will be required from impact assessments. This includes the impact on outcomes of patients with drug-sensitive and drug-resistant disease, as well as the impact on programmatic outcomes at facility, district, regional and national levels. Demonstration of mortality reduction and reductions in TB transmission would go a long way to providing strong justification for full scale-up. These data will need to be incorporated in thorough cost–effectiveness assessments of the assay.
Future perspective
To eliminate TB as a public health problem (as defined by a TB incidence of less than one per million population worldwide) by 2050, TB incidence rates will need to decrease by an average of 16% per year for the next 40 years [41]. No single intervention will achieve this, but modeling studies have shown that new diagnostic tests could make a substantial impact. Implementation of a nucleic acid amplification test globally was estimated to reduce TB incidence by 28% overall by 2050 [42]. However, the impact of even the most promising new diagnostic test will be highly dependent upon the system within which it is used and, more specifically, whether it reaches the patients who may benefit from it. We are at a critical juncture in that a diagnostic tool with excellent performance characteristics has been developed, but there is a crucial need for appropriate implementation to fully realize the benefits. In the immediate future, operational and implementation researchers need to quickly identify and respond to all the issues associated with the implementation and scale-up of this new technology. TB programs must adapt their systems to use this technology, increasing access to patients while also focusing on the basics of curative TB treatment so that increased case-finding will translate into more patients cured. Moreover, there is a critical need for increased capacity to diagnose MDR-TB to be matched by increased capacity to effectively treat and cure diagnosed cases. Patient advocates and activists should hold everyone accountable and drive demand for better TB programmes that include use of new tools such as the Xpert MTB/RIF assay. Impact assessment of initial scale-up in high burden countries over the next few years will quickly give an indication as to whether this technology truly does hold promise for delivering improvements to TB control in the 21st century.
| Study† | Description | Ref. |
|---|---|---|
| Development of prototype assay | ||
| Piatek et al. | Development of the molecular beacon-based assay targeting the rpoB region of Mycobacterium tuberculosis | [5] |
| Piatek et al. | Assessment of prototype multiwell molecular beacon assay for detection of rifampicin and isoniazid resistance in lysates of clinical M. tuberculosis isolates from New York and from Spain | [6] |
| El-Hajj et al. | Further development of a single-well molecular beacon assay for detection of rifampicin resistance and evaluation using 148 clinical isolates of M. tuberculosis | [7] |
| Laboratory based studies of analytic performance & biosafety of Xpert® MTB/RIF | ||
| Helb et al. | First description of the Xpert MTB/RIF assay reporting on the sterilizing activity of the sample reagent; the determination of the limits of detection for genomic DNA and for M. tuberculosis spiked into sputum samples; assessment of cross reactivity with nontuberculous mycobacterial species; a small clinical validation study of archived sputum samples from Vietnam and Uganda (n = 132) | [8] |
| Blakemore et al. | Study of the dynamic range of the assay; the analytic sensitivity and specificity using DNA from phylogenetically and geographically diverse clinical isolates of M. tuberculosis (n = 79) including rifampicin-resistant isolates (n = 37) and a range of bacterial, fungal and viral clinical isolates (n = 89); the ability to detect rpoB mutations in varying mixtures of wild-type and mutant DNA; the impact of contamination with amplicons from the MTBDR plus assay | [9] |
| Banada et al. | Study examining the sterilizing activity of sample reagent and assessing viable bioaerosol generation during the manual and automated phases of the assay compared with smear microscopy | [10] |
| FIND multicountry evaluations | ||
| Boehme et al. | A multicountry (South Africa, Peru, Azerbaijan and India) laboratory validation of the assay to determine the sensitivity and specificity when testing clinical samples from 1730 patients suspected of having TB or multidrug resistant-TB | [11] |
| Boehme et al. | A multicenter (South Africa, Peru, India, Azerbaijan and the Philippines) implementation study to assess the operational feasibility, accuracy and impact on time to diagnosis and time to start of TB treatment when the assay is used at the district or subdistrict level | [12] |
| Studies of diagnostic accuracy in clinical samples in the USA, Europe & India | ||
| Marlowe et al. | Laboratory-based assessment of routine respiratory samples (n = 217) in the USA | [24] |
| Armand et al. | Laboratory-based comparison of the sensitivity of the assay with that of an in-house IS6110-based real time-PCR assay using routine respiratory and nonrespiratory samples (n = 117) in France | [25] |
| Moure et al. | Laboratory-based assessment of diagnostic accuracy when testing respiratory and nonrespiratory (n = 85) routine clinical samples in Spain | [26] |
| Hillemann et al. | Laboratory-based assessment of diagnostic accuracy when testing nonrespiratory routine clinical samples (n = 521; urine, stool, cerebrospinal fluid, pleural fluid, gastric aspirate and tissue) in Germany | [31] |
| Bowles et al. | Laboratory-based assessment of diagnostic accuracy when testing fresh and frozen samples (n = 89, predominantly sputum) in The Netherlands | [27] |
| Vadwai et al. | Assessment of diagnostic accuracy in prospectively recruited hospital in-patients (n = 546) with suspected extrapulmonary TB in India | [33] |
| Causse et al. | Laboratory-based comparison of the diagnostic accuracy of Xpert MTB/RIF with Cobas® TaqMan® MTB in 340 nonrespiratory samples | [32] |
| Ioannidis et al. | Laboratory-based assessment of diagnostic accuracy when testing pulmonary and extrapulmonary samples (n = 121) from Greece | [43] |
| Studies of the investigation of TB suspects in African settings | ||
| Nicol et al. | Prospective study from South Africa of pediatric in-patients (n = 452, median age 19 months) with suspected TB | [28] |
| Theron et al. | A retrospective assessment of assay sensitivity and specificity using frozen sputum samples from TB suspects (n = 496) in a high HIV-prevalence setting in South Africa | [30] |
| Rachow et al. | A retrospective assessment of sensitivity and specificity using frozen sputum samples from TB suspects (n = 292) in Tanzania | [105] |
| Van Rie et al. | Initial report from a study evaluating the role of the assay when applied to routine fine needle aspirates obtained from HIV-infected TB suspects (n = 101) with lymphadenopathy in South Africa | [34] |
| Study of routine screening for HIV-associated TB | ||
| Lawn et al. | A prospective study of HIV-infected patients (n = 515) with advanced immunodeficiency being systematically screened prior to antiretroviral therapy in South Africa | [35] |
| Study of utility in TB clinical trials | ||
| Friedrich et al. | A study demonstrating the utility of the assay for rapid enrollment of patients into clinical TB trials | [44] |
| 2011 | 2012 | 2014 | |
|---|---|---|---|
| Estimated global demand (cartridge numbers) | >600,000 | >1,700,000 | >3,700,000 |
| Negotiated price (US$) | 18.40 | 14.00 | 10.72 |
| Average reduction over average European price (€50) | 75% | 79% | 84% |
Executive summary
The need for new TB diagnostics
▪ The lack of simple, accurate and rapid diagnostics is a major hindrance to TB control, especially in high-burden settings where sputum smear microscopy is the mainstay of diagnosis.
The Xpert® MTB/RIF assay
▪ Mutations in the 81 bp core of the rpoB gene of Mycobacterium tuberculosis account for >95% of rifampicin resistance and flanking sequences are specific to the organism.
▪ The PCR-based assay is almost completely automated, being conducted with a multichambered plastic cartridge that is preloaded with all the necessary reagents. This is inserted within the GeneXpert diagnostic platform, which reports the presence of M. tuberculosis and rifampicin resistance.
Laboratory-based evaluation
▪ The limit of detection of the assay (95% sensitivity) is 131 CFU/ml of M. tuberculosis in sputum; it shows no cross-reactivity with non-tuberculous mycobacteria or with a range of respiratory tract pathogens.
▪ The assay showed 100% specificity and sensitivity for detection of a wide range of rpoB mutations.
▪ The assay was not found to generate infectious bioaerosols.
Evaluation in clinical populations
▪ Testing a single sputum sample detects 98–100% of sputum smear-positive pulmonary TB and between 57% and 83% of sputum smear-negative disease with very high specificity in adults. The assay also doubles case detection in children when compared to smear microscopy.
▪ When used to screen HIV-infected patients before starting antiretroviral therapy, Xpert increased case detection compared to smear microscopy by 45% (28% vs 73%).
▪ Initial studies of a range of nonrespiratory samples from sites of extrapulmonary TB have reported sensitivities of 53–95% for TB diagnosis.
▪ The first FIND study found a sensitivity (99.1%) and specificity (100%) for detection of rifampicin resistance, but subsequent studies have discovered numbers of cases of false-positive rifampicin resistance.
Implementation
▪ A large-scale evaluation found that Xpert was successfully implemented at the district and sub-district level in urban settings, accelerating diagnosis and start of TB treatment and reducing the proportion of untreated disease.
WHO endorsement, costs & implementation
▪ In December 2010 the WHO endorsed use of this assay as the initial diagnostic test in individuals suspected of having MDR-TB or HIV-associated TB (strong recommendation).
▪ WHO also endorsed use of the test as a follow-on to smear microscopy in settings where MDR-TB and HIV-associated TB were less of a concern (conditional recommendation).
▪ Phased implementation from 2011 onwards is recommended, leading to full scale-up thereafter. The cost of cartridges will decrease over time.
Unresolved issues
▪ A huge number of issues remain to be addressed with regards to operational implementation, cost–benefit and cost–effectiveness, and solutions to the problem of false-positive rifampicin resistance results.
Financial & competing interests disclosure
SD Lawn and MP Nicol are both funded by the Wellcome Trust, London, UK, and MP Nicol has also received funding from EDCTP and the NIH (USA). MP Nicol has received funding from FIND to conduct evaluation and implementation studies of Xpert® in South Africa. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Papers of special note have been highlighted as: ▪ of interest
Bibliography
- 1 Lawn SD, Zumla AI. Tuberculosis. Lancet378,57–72 (2011).
- 2 Gandhi NR, Nunn P, Dheda K et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet375,1830–1843 (2010).
- 3 Pai M, Minion J, Sohn H, Zwerling A, Perkins MD. Novel and improved technologies for tuberculosis diagnosis: progress and challenges. Clin. Chest Med.30,701–716 (2009).
- 4 Small PM, Pai M. Tuberculosis diagnosis – time for a game change. N. Engl. J. Med.363,1070–1071 (2010).
- 5 Piatek AS, Tyagi S, Pol AC et al. Molecular beacon sequence analysis for detecting drug resistance in mycobacterium tuberculosis. Nat. Biotechnol.16,359–363 (1998).
- 6 Piatek AS, Telenti A, Murray MR et al. Genotypic analysis of mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother.44,103–110 (2000).
- 7 El-Hajj HH, Marras SA, Tyagi S, Kramer FR, Alland D. Detection of rifampin resistance in mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol.39,4131–4137 (2001).
- 8 Helb D, Jones M, Story E et al. Rapid detection of mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol.48,229–237 (2010).▪ First description of the Xpert® MTB/RIF assay reporting on the analytical laboratory performance and a proof of concept clinical validation using frozen sputum samples.
- 9 Blakemore R, Story E, Helb D et al. Evaluation of the analytical performance of the Xpert(R) MTB/RIF assay. J. Clin. Microbiol.48,249–251 (2010).
- 10 Banada PP, Sivasubramani SK, Blakemore R et al. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J. Clin. Microbiol.48,3551–3557 (2010).
- 11 Boehme CC, Nabeta P, Hillemann D et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med.363,1005–1015 (2010).▪ The seminal multicountry laboratory validation of the assay demonstrating for the first time the excellent diagnostic accuracy among prospectively recruited TB suspects.
- 12 Boehme CC, Nicol MP, Nabeta P et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet377,1495–1505 (2011).▪ The first large-scale field implementation study demonstrating feasibility and impact at the district and sub-district level.
- 13 Greco S, Girardi E, Navarra A, Saltini C. Current evidence on diagnostic accuracy of commercially based nucleic acid amplification tests for the diagnosis of pulmonary tuberculosis. Thorax61,783–790 (2006).
- 14 Ling DI, Flores LL, Riley LW, Pai M. Commercial nucleic-acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta-analysis and meta-regression. PLoS ONE3,e1536, (2008).
- 15 Pai M, Flores LL, Hubbard A, Riley LW, Colford JM Jr. Nucleic acid amplification tests in the diagnosis of tuberculous pleuritis: a systematic review and meta-analysis. BMC Infect. Dis.4,6 (2004).
- 16 Pai M, Flores LL, Pai N, Hubbard A, Riley LW, Colford JM Jr. Diagnostic accuracy of nucleic acid amplification tests for tuberculous meningitis: a systematic review and meta-analysis. Lancet Infect. Dis.3,633–643 (2003).
- 17 Lawn SD, Churchyard G. Epidemiology of HIV-associated tuberculosis. Curr. Opin. HIV AIDS4,325–333 (2009).
- 18 Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev.8,496–514 (1995).
- 19 Sreevatsan S, Pan X, Stockbauer KE et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl Acad. Sci. USA94,9869–9874 (1997).
- 20 Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol.14,303–308 (1996).
- 21 Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol.16,49–53 (1998).
- 22 Ulrich MP, Christensen DR, Coyne SR et al. Evaluation of the cepheid GeneXpert system for detecting bacillus anthracis. J. Appl. Microbiol.100,1011–1016 (2006).
- 23 No authors listed. Diagnostic Standards and Classification of Tuberculosis in Adults and Children. This official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999. This statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. Am. J. Respir. Crit. Care Med.161,1376–1395 (2000).
- 24 Marlowe EM, Novak-Weekley SM, Cumpio J et al. Evaluation of the cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J. Clin. Microbiol.49,1621–1623 (2011).
- 25 Armand S, Vanhuls P, Delcroix G, Courcol R, Lemaitre N. Comparison of the Xpert MTB/RIF test with an IS6110-TaqMan real-time PCR assay for direct detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens J. Clin. Microbiol.49,1772–1776 (2011).
- 26 Moure R, Munoz L, Torres M, Santin M, Martin R, Alcaide F. Rapid detection of Mycobacterium tuberculosis complex and rifampin resistance in smear-negative clinical samples by use of an integrated real-time PCR method. J. Clin. Microbiol.49,1137–1139 (2011).
- 27 Bowles EC, Freyee B, van Ingen J, Mulder B, Boeree MJ, van SD. Xpert MTB/RIF(R), a novel automated polymerase chain reaction-based tool for the diagnosis of tuberculosis. Int. J. Tuberc. Lung Dis.15,988–989 (2011).
- 28 Nicol MP, Workman L, Isaacs W et al. A descriptive study of the accuracy of the Xpert MTB/RIF test for diagnosis of pulmonary tuberculosis in hospitalized children in a high HIV prevalence area. Lancet Infect. Dis. doi:10.1016/S1473-3099(11)70167-0 (2011) (Epub ahead of print).▪ The first evaluation of the diagnostic accuracy of the Xpert MTB/RIF test in children.
- 29 Hatherill M, Hanslo M, Hawkridge T et al. Structured approaches for the screening and diagnosis of childhood tuberculosis in a high prevalence region of South Africa. Bull. World Health Organ.88,312–320 (2010).
- 30 Theron G, Peter J, van Zyl-Smit R et al. Evaluation of the Xpert(R) MTB/RIF assay for the diagnosis of pulmonary tuberculosis in a high HIV prevalence setting. Am. J. Respir. Crit. Care Med. (2011).
- 31 Hillemann D, Rusch-Gerdes S, Boehme C, Richter E. Rapid molecular detection of extrapulmonary tuberculosis by the automated GeneXpert MTB/RIF system. J. Clin. Microbiol.49,1202–1205 (2011).
- 32 Causse M, Ruiz P, Juan Bautista GA, Casal M. Comparison of two molecular methods for the rapid diagnosis of extrapulmonary tuberculosis. J. Clin. Microbiol.49(8),3065–3067 (2011).
- 33 Vadwai V, Boehme C, Nabeta P, Shetty A, Alland D, Rodrigues C. Xpert MTB/RIF, a new pillar in the diagnosis of extrapulmonary tuberculosis? J. Clin. Microbiol.49(7),2540–2545 (2011).
- 34 Van Rie A, Menezes C, Scott L et al. High yield, sensitivity and specificity of Xpert MTB/RIF for M. tuberculosis detection in fine needle aspirates from HIV-infected TB suspects. Program and abstracts of the 18th Conference on Retroviruses and Opportunistic Infections. Boston, MA, USA. (2011) (Abstract #879).
- 35 Lawn SD, Brooks SV, Kranzer K et al. Screening for HIV-associated tuberculosis and rifampicin resistance before antiretroviral therapy. A prospective study. PLoS Med.8(7),e1001067 (2011).▪ The first evaluation of the assay used as a routine screening test among HIV-infected patients with advanced immunodeficiency before starting antiretroviral therapy.
- 36 Lawn SD, Wood R. Tuberculosis in antiretroviral treatment services in resource-limited settings: addressing the challenges of screening and diagnosis. J. Infect. Dis. (2011) (In Press).
- 37 Bassett IV, Wang B, Chetty S et al. Intensive tuberculosis screening for HIV-infected patients starting antiretroviral therapy in Durban, South Africa. Clin. Infect. Dis.51,823–829 (2010).
- 38 Lawn SD, Edwards DJ, Kranzer K, Vogt M, Bekker LG, Wood R. Urine lipoarabinomannan assay for tuberculosis screening before antiretroviral therapy diagnostic yield and association with immune reconstitution disease. AIDS23,1875–1880 (2009).
- 39 Lawn SD, Wood R. Tuberculosis screening in patients starting antiretroviral therapy in sub-Saharan Africa: stretching diagnostics to the limits. Clin. Infect. Dis.52,276–277 (2011).
- 40 Cox HS, McDermid C, Azevedo V et al. Epidemic levels of drug resistant tuberculosis (MDR and XDR-TB) in a high HIV prevalence setting in Khayelitsha, South Africa. PLoS ONE5,e13901 (2010).
- 41 Lonnroth K, Castro KG, Chakaya JM et al. Tuberculosis control and elimination 2010–50: cure, care, and social development. Lancet375,1814–1829 (2010).
- 42 Abu-Raddad LJ, Sabatelli L, Achterberg JT et al. Epidemiological benefits of more-effective tuberculosis vaccines, drugs, and diagnostics. Proc. Natl Acad. Sci. USA106,13980–13985 (2009).
- 43 Ioannidis P, Papaventsis D, Karabela S et al. Cepheid GeneXpert MTB/RIF assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with high clinical suspicion of TB and smear negative microscopy. J. Clin. Microbiol.49(8),3068–3070 (2011).
- 44 Friedrich SO, Venter A, Kayigire XA, Dawson R, Donald PR, Diacon AH. Xpert MTB/RIF and genotype MTBDRplus for patient selection for a tuberculosis clinical trial (JCM00138-11). J. Clin. Microbiol.49(8),2827–2831 (2011).
- 101 WHO. Global tuberculosis control 2010. Geneva: WHO, 2010 www.who.int/tb/publications/global_report/2010/en/index.html
- 102 WHO. Multidrug and extensively drug-resistant TB (M/XDR-TB). 2010 global report on surveillance and response. WHO, Geneva, 2010. WHO/HTM/TB/2010.3 http://whqlibdoc.who.int/publications/2010/9789241599191_eng.pdf (Accessed 18th August 2010)
- 103 WHO. Molecular line probe assays for rapid screening of patients at risk of multidrug-resistant tuberculosis (MDR-TB). Policy statement June 2008 www.who.int/entity/tb/dots/laboratory/lpa_policy.pdf
- 104 Cepheid Systems www.cepheid.com/systems-and-software/systems-overview
- 105 Rachow A, Mtafya B, Rojas-Ponce G et al. Detection of Mycobacterium tuberculosis using the Cepheid Xpert MTB/RIF assay – a clinical validation study from Tanzania. 41st Union World Conferenec on Lung Health. Berlin, Germany, 11–15 November 2010 Abstract session 01 http://uwclh.conference2web.com/content/667 (Accessed 19th April 2011)
- 106 Nicol M. Algorithms for referral to rapid diagnostics for MDR-TB: preliminary outcomes. 41st Union World Conference on Lung Health. International Union Against Tuberculosis and Lung Disease. Berlin, Germany, November 2010. Session #10 http://uwclh.conference2web.com/content/227 (Accessed 2nd February 2011
- 107 WHO. Rapid implementation of the Xpert MTB/RIF diagnostic test: technical and operational ‘How to’; practical considerations. WHO, Geneva. 2011 http://whqlibdoc.who.int/publications/2011/9789241501569_eng.pdf (Accessed 28th June 2011)
- 108 WHO. Tuberculosis diagnostics automated DNA test. WHO endorsement and recommendations. World Health Organization, Geneva, 2010 www.who.int/tb/features_archive/xpert_factsheet.pdf
- 109 WHO. Roadmap for rolling out Xpert MTB/RIF for rapid diagnosis of TB and MDR-TB. World Health Organization, Geneva, December 2010 www.who.int/tb/laboratory/roadmap_xpert_mtb_rif_rev23dec2010.pdf (Accessed 8th December 2010)
































